|
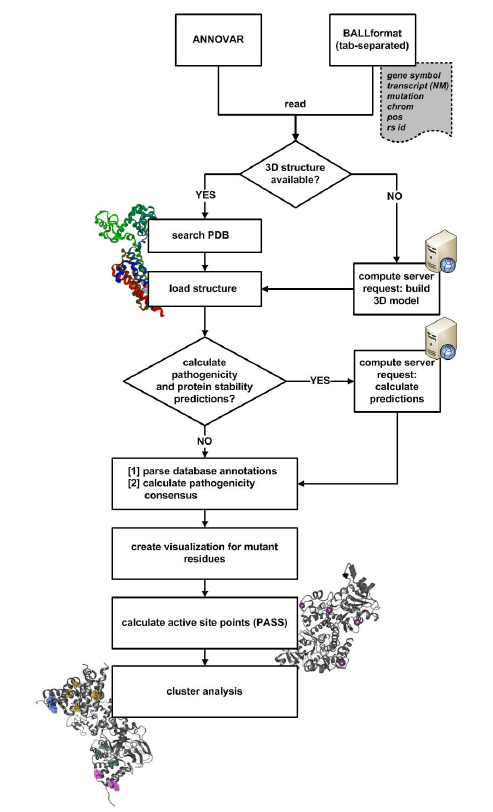
BALL-SNP is a software tool based on the Biochemical Algorithms Library (BALL), a molecular modeling framework, providing robust and sophisticated algorithms on structural bioinformatics. BALL-SNP enables the assessment of multiple non-synonymous single nucleotide polymorphisms (nsSNPs) in a single protein by visualizing the mutated residues within the wild type structure, collecting available pathogenicity information from different databases and predicting binding pockets as well as protein stability changes. Based on the generated information and the three-dimensional visualization, a user can assume whether the amino acid substitutions can have a quantitative effect due to mutual interaction or have an influence on binding and stability. Input formatsAn input file containing nsSNP data can be loaded via the
When an input file with nsSNP information is opened, BALL-SNP processes the following pipeline:
Examples
Exemplary data and its analyses can be found here.
Demonstration Videos
For a better understanding of BALL-SNP and its functionality, we will provide videos showing the above described examples.
Comming soon….
Description of the main windowHow to …InstallationDetails how to install BALL-SNP:here |

 button in the menu.
button in the menu.