|
The structure window gives a hierarchical overview of the loaded molecules.
 Highlighting
By clicking with the left mouse button on the names of molecular entities (system, protein, chain),
users can highlight these.  Selections (or how to find an atom)Left of the names of the molecular entities are checkboxes, which show the user which molecular entities are
currently selected (picked). The selected objects become colored in yellow in the 3D View. This can be used to find an atom in the three-dimensional structure of its molecule. 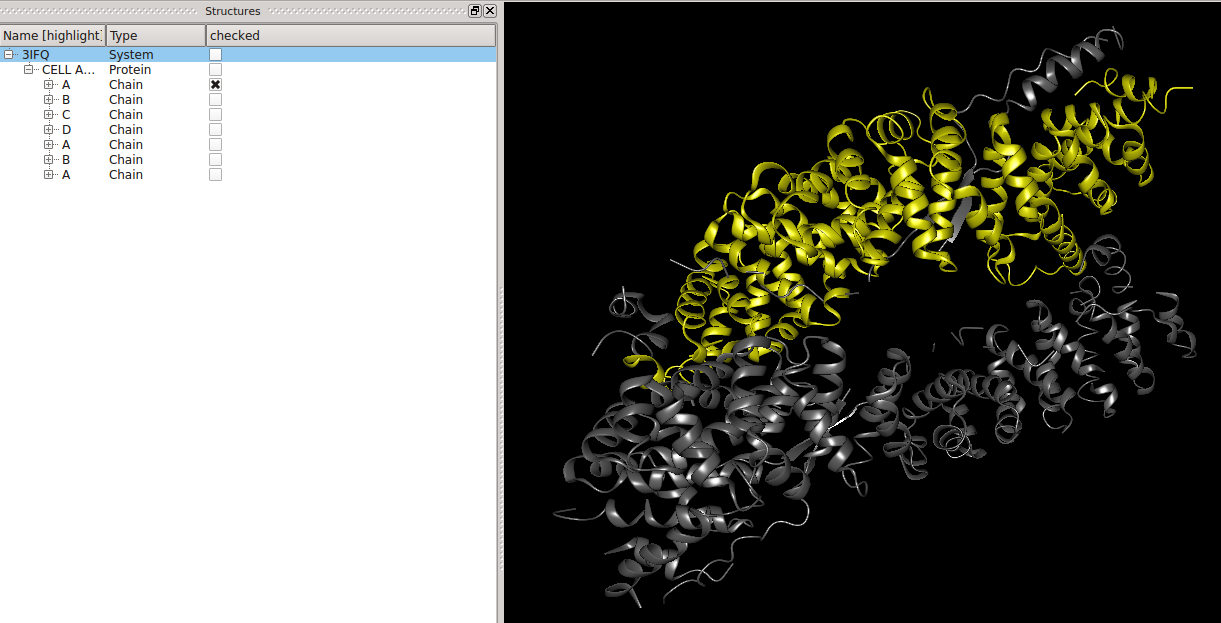 BALL expressionsAt the bottom of the structure window resides a text field.
In this text field, the user can enter expressions to select molecular entities. To apply the typed expression, either press the Return key or the “Select” button. The “Clear” button clears the current selection. Possible predicates are:
Reset BALL-SNP
Before loading a new input file containing SNP data, BALL-SNP has to be reset and the currently loaded SNP data and the generated information deleted. To do so, the loaded molecular entities, namely the system or protein, has to be highlighted and deleted with either str + entf. This is necessary, since BALL-SNP currently is not able to load more than one input file at once. However, mixing up different genes and the 3D structures of the encoded proteins is not very useful, in your point of view.
Back to BALL-SNP tutorial main page.
|

 .
.